gpt3_radreports
Translate radiology reports from clinical to lay text
Project maintained by DrHughHarvey Hosted on GitHub Pages — Theme by mattgraham
Introduction
Radiologists use a specific ontology for describing imaging findings in their reports. This language is esoteric and poorly understood by lay readers. With the advent of few-shot learning natural language models such as GPT3, it is now possible to train billion parameter networks to convert input text into output text in a stylised fashion, using only a few training examples.
The aim of this project is to build a simple web interface using theOpenAI GPT-3 APIwhere the input prompt can be any clinical radiology report and the resulting output is an understandable lay version.
Context
Human translation of standard radiology reports into lay terms would be extremely time consuming, hence this is never done in routine clinical practice. Instead radiology reports are read by clinicians, and then summarised either verbally or via a clinic letter to their patient. Current radiological practice therefore is non patient-centric in that the end-user of the service neither receives their report, nor could understand their report if they did receive it.
An example of the esoteric nature of radiology reports is:
There are several hyerechoic calcific foci within the gallbladder with
significant posterior acoustic shadowing in keeping with cholecystolithiasis
where the lay version may be translated as:
There are several dense objects typical of gallstones within the gallbladder
In keeping with the recent trend towards a more patient-focussed model of healthcare, and increasing appetite for digital tools to enable and empower both physicians and patients, this project aims to bridge the ‘language’ barrier that lies between radiologists and patients by using the powerful natural language capabilities of GPT3.
The tool is loaded in three parts:
- Set up the API configuration in a virtual environment
- Load a web interface to allow user input and visualisation of the output
- ‘Prime’ GPT3 for the specific task using examples
How to run gpt3_radreports
Set up the API configuration in a virtual environment
The code in this repo is based onShreyaShankar's gpt3-sandbox-> here
I also recommend this youtube video to explain various elements of this sandbox.
The following has only been tested on MacOS Catalina
- Obtain OpenAI API key - there is a waiting list, so be patient!
- Run
Terminal - Install python3
- Install yarn
Clone this repository, and in Terminal install the Python virtual environnment package, create a new virtual environment called ‘gpt3’ and activate it:
pip3 install virtualenv
python3 -m venv gpt3
source gpt3/bin/activate
Go to the root folder of the repository:
cd /your/path/to/the/root/gpt3
Install the requirements package
pip install -r api/requirements.txt
Open openai.cfg in TextEdit and exchangeINSERT_OPEN_AI_KEY_HEREfor your OpenAI secret key.
Link your gpt3 virtual environment to the config file containing the API key.
export OPENAI_CONFIG=/YOUR/PATH/TO/THE/CONFIG/FILE/api/openai.cfg
Install yarn (this will create a file in your repository called yarn.lock which you should not delete)
yarn install
Load the web interface to allow user input and visualisation of the output
Onceyarnis installed and theopeanai.cfgkey has been linked (as above), you can load the web app.
Load the web app by running the example script included in the gpt3_radreports repository:
python /examples/run_radiologyexamples.py
Your browser will open a new tab which should display the following:
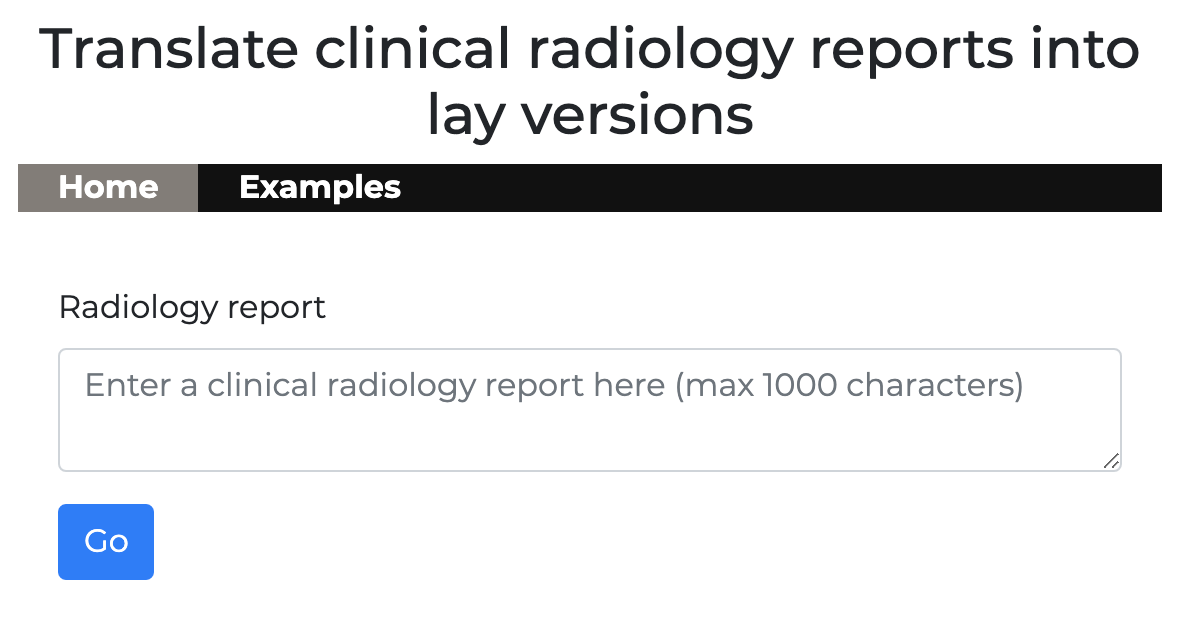
There are two tabs in the web app
- Home - for querying GPT3 via the API
- Examples - for priming GPT3
‘Prime’ GPT3 for the specific task using examples
GPT3 can only ‘solve’ the task if it has been shown a few examples (known as ‘few-shot learning’), so we need to create some cases for it to learn from - this is called priming. This is done using a simple python script, and the full code is ingpt3/examples/run_radiologyreports.pywhich loads the web app.
To be able to send examples to GPT3 via our API we created a ‘GPT object’ with the following parameters:
*engine - this will be davinci which is the current version of the GPT3 engine
*temperature - this is a setting between 0-1 which tells GPT3 how accurate or creative to be - 0 being very accurate, 1 being more creative
*max_tokens - how many characters do we want the output to be?
gpt = GPT(engine="davinci", temperature=0.2, max_tokens=1000)
Next we created some examples we want to use to prime GPT3.
e.g:
gpt.add_example(Example("There are several hyerechoic calcific foci within the gallbladder with significant posterior acoustic shadowing in keeping with cholecystolithiasis", "There are several dense objects typical of gallstones within the gallbladder"))
When you load the web app and visit the examples tab, you will see the six examples I have selected to prime GPT3. You can add more of your own by clicking the ‘add example’ button at the bottom.
Stop the web app at any time by running command-c in Terminal.
These are the 6 pre-selected examples. You can prime GPT3 using different examples, and take note of how the outputs change.
Example 1
*input*
There are several hyerechoic calcific foci within the gallbladder with significant posterior acoustic shadowing in keeping with cholecystolithiasis.
*output*
There are several dense objects typical of gallstones within the gallbladder.
Example 2
*input*
The lungs and pleura are clear and the cardiomedistinal silhouette is unremarkable. No rib lesions.
*output*
Normal chest X-ray.
Example 3
*input*
No mass, haemorrhage or hydrocephalus. Basal ganglia and posterior fossa structures are normal. No established major vessel vascular territory infarct. No intra or extra axial collection. The basal cisterns and foramen magnum are patent. The air cells of the petrous temporal bone are non-opacified. No fracture demonstrated.
*output*
Normal CT head.
Example 4
*input*
Low signal intensity foci seen in the posterior aspect of the peripheral zone at the level of the mid gland toward the apex measuring 0.9 cm on the left and 0.9 cm on the right with associated early enhancement and possible restricted diffusion. Findings are concerning for neoplastic foci.
*output*
There are a couple of areas which could be cancerous in the outer part of the prostate.
Example 5
*input*
Prominent bilobed paramedial extra-axial mass along the convexity centered at the level of the posterior frontal and anterior parietal lobes with prominent posterior dural tail and occlusion of the adjacent superior sagittal sinus. Prominent surrounding reactive edema, left greater than right. Mild lateral shift but no herniation. Smaller extraaxial mass overlying the right mid temporal lobe.
*output*
There is one large right-sided mass near the front of the brain, and another smaller one on the same side. These are likely to be tumours, with signs of brain swelling.
Example 6
*input*
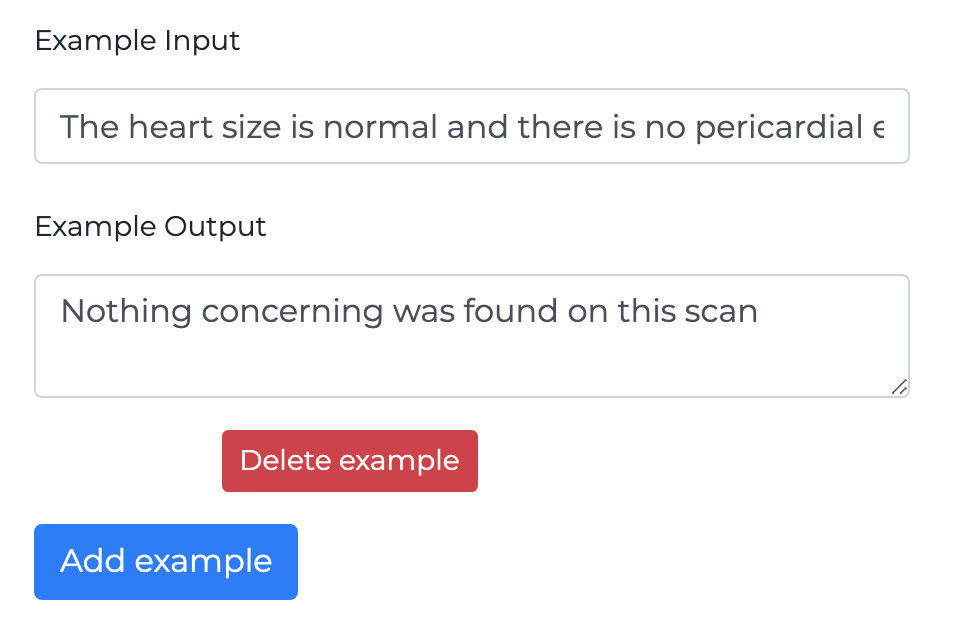
The heart size is normal and there is no pericardial effusion. The aorta and great vessels are normal in caliber. The central pulmonary arteries are patent with no evidence of embolus. There is no significant mediastinal, hilar, or axillary lymphadenopathy. The trachea and mainstem bronchi are patent. The esophagus is normal in course and caliber. The lungs are clear with no infiltrates, effusions, or masses. There is no pneumothorax. Scans through the upper abdomen are unremarkable. The osseous structures in the chest are intact.
*output*
Nothing concerning was found.